| Research work: 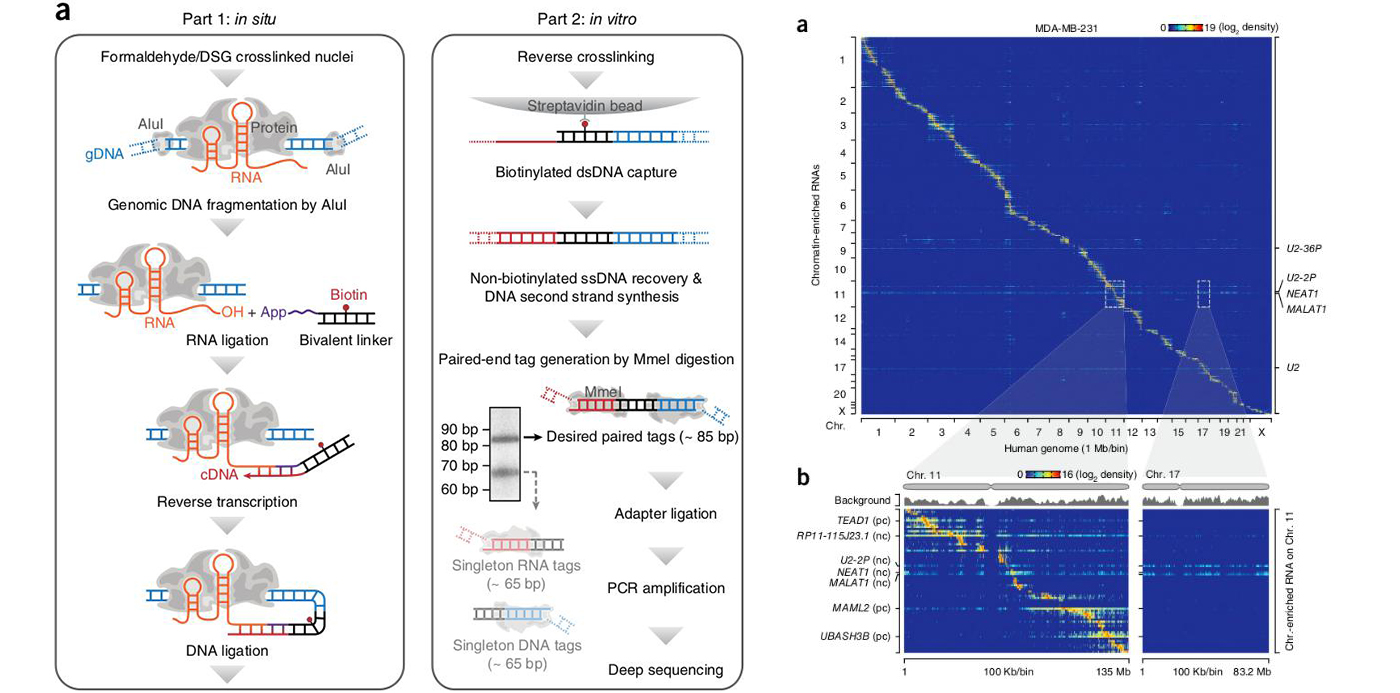
We developed a method to capture global RNA interactions on chromatin in situ by deep sequencing (GRID-seq) in fixed permeabilized nuclei that allows identification of the entire repertoire of chromatin-associated RNAs in an unbiased manner(Nature biotech. 2017)
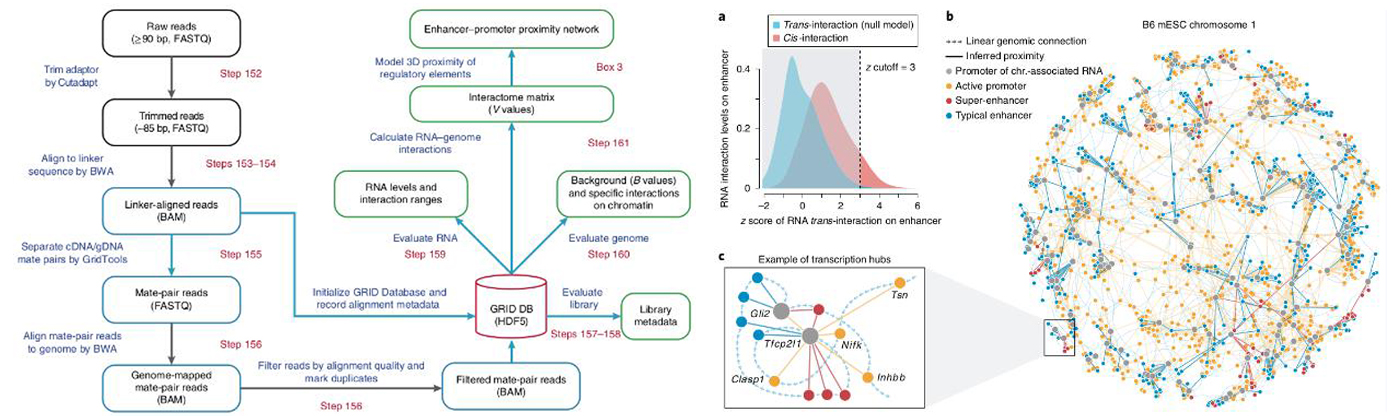
Enhanced the utility of GRID-seq, we developed a pipeline for integrative analysis on multi-omics data, called GridTools, into which key steps such as modeling of enhancer-promoter connectivity and inference of genomic element proximity are integrated.(Nature prot. 2019)
| Plain english:
Our research focuses on the fundamental questions about how cell identity is determined, maintained, actively regulated, and how single cells are systematically aging and regeneration in the context of heterogenic microenvironment of the tissue or organ. Combining computational methodologies with high-throughput sequencing technologies, our researches specifically focus on the characterization, organization, function and regulatory mechanism of single cells in the multicellular microenvironment, in various levels of functional genomics, epigenetics, and chromatin architectures. Selected publications: - Bing Zhou, Xiao Li, Daji Luo, Do-Hwan Lim, Yu Zhou, and Xiang-Dong Fu. GRID-seq for comprehensive analysis of global RNA-chromatin interactions. Nature Protocols, 2019.
- Peng Liu^, Shuaibin Zhang^, Bing Zhou^, Xi Luo, Xiaofeng Zhou, Bin Cai, Yinhua Jin, De Niu, Jinxing Lin, Xiaofeng Cao, and Jingbo Jin. The histone H3K4 Demethylase JMJ16 represses leaf senescence in Arabidopsis. The Plant Cell, 2019.
- Xiao Li^, Bing Zhou^, Liang Chen, Lan-Tao Gou, Hairi Li, Xiang-Dong Fu. GRID-seq reveals the global RNA-chromatin interactome. Nature biotechnology, 2017.
- Jinsong Qiu^, Bing Zhou^, Felicitas Thol, Yu Zhou, Liang Chen, Changwei Shao, Christopher DeBoever, Jiayi Hou, Hairi Li, Anuhar Chaturvedi, Arnold Ganser, Rafael Bejar, Dong-Er Zhang, Xiang-Dong Fu, Michael Heuser. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA, 2016.
- Xia Cui^, Falong Lu^, Qi Qiu^, Bing Zhou^, Lianfeng Gu, Shuaibin Zhang, Yanyuan Kang, Xiekui Cui, Xuan Ma, Qingqing Yao, Jinbiao Ma, Xiaoyu Zhang, Xiaofeng Cao. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nature genetics, 2016.
- Shuaibin Zhang^, Bing Zhou^, Yanyuan kang, Xia Cui, Ao Liu, Angelique Deleris, Maxim V. C. Greenberg, Xiekui Cui, Qi Qiu, Falong Lu, James A. Wohlschlegel, Steven E. Jacobsen and Xiaofeng Cao. C-terminal domains of histone demethylase JMJ14 interact with a pair of NAC transcription factors to mediate specific chromatin association. Cell Discovery, 2015.
- Bing Zhou^, Liu Yang^, Shoufeng Li, Jialiang Huang, Haiyang Chen, Lei Hou, Jinbo Wang, Christopher D. Green, Zhen Yan, Xun Huang, Matt Kaeberlein, Li Zhu, Huasheng Xiao, Yong Liu, and Jing-Dong J. Han. Midlife gene expressions identify modulators of aging through dietary interventions. PNAS, 2012.
- Hong Yu^, Shanshan Zhu^, Bing Zhou^, Huiling Xue, and Jing-Dong Jackie Han. Inferring causal relationships among different histone modifications and gene expression. Genome Research, 2008.
|

