 | Jing QU Stem Cells and Aging
Email: qujing@ioz.ac.cn
Staff: Jing Qu, Yanxia Ye, Zhejun Ji, Qun Chu, Ruijun Bai, Shangyi Qiao
Students: Zehua Wang, Yandong Zheng, Min Wang, Chengyu Liu, Yixin Zhang, Baohu Zhang, Mingheng Li, Shuaiqi Fang, Tianxiang Zhang, Dingyi Liu, Chunyu Jiao
| | |
| Introduction Jing Qu earned her B.S. from Lanzhou University in 2002, and then the doctor degree from the Institute of Biophysics, CAS in 2007. From 2007 to 2012, she worked as a Postdoctoral Fellow at the Del E. Web Neuroscience, Aging, and Stem Cell Research Center at the Sanford/Burnham Medical Research Institute, and then as a Research Associate in the Gene Expression Laboratory at the Salk Institute for Biological Studies. Since 2014, she established the “ Stem Cell and Aging” Lababory at the Institute of Zoology, CAS. She is now the Chair of the Aging Genetics Elites of the Genetics Society of China, and also a member of Scientific Program Committee of ISSCR. She received the Chinese Young Women in Science Fellowship for her contributions to aging research. Research work 1) Development of novel platform for aging research Based on the non-human-primate organ aging research system, combined with in vitro aging models of human stem cells and their derivative cells, organoids, providing a novel platform for aging research is a prerequisite for the mechanism exploration for human aging. Our recent related work includes, establishing the first multi-tissue single-cell mammalian transcriptomic atlas undergoing aging and aging interventions and revealed new molecular mechanisms in delaying aging; drawing the first high-resolution single-cell transcriptome maps of primate organs aging and reveal the key drivers and biomarkers of aging, Development of an aging database, Aging Atlas, to integrate multi-species aging-related data sets, with commitment to the convergence, open sharing and cross-research of aging-related big data (Figure 1). 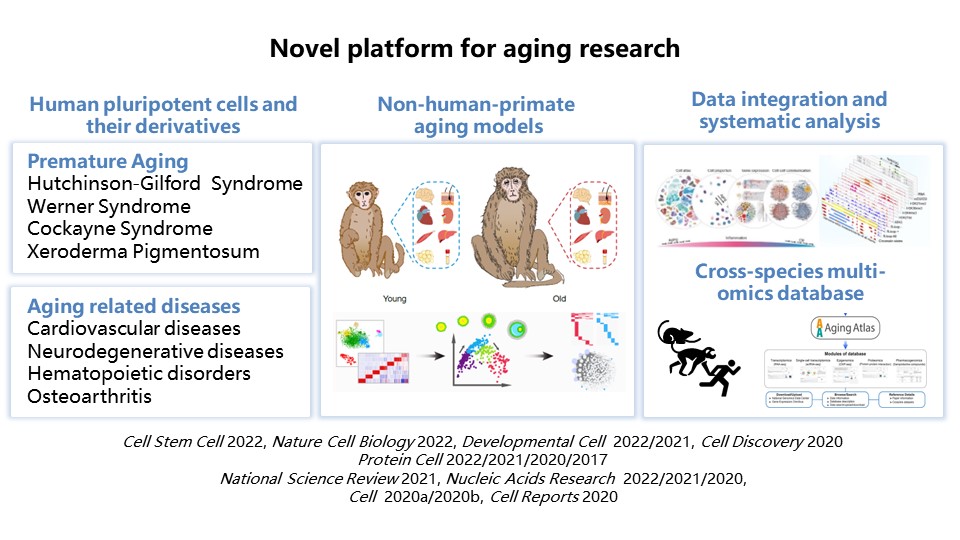
Figure 1. Novel platform for aging research
2) Systematic investigation of regulatory mechanisms underlying aging Aiming at the heterogeneity, nonlinearity, and spatiotemporal specificity of organ aging, we systematically investigated the convergent and specific aging features for different organs and cell types, to explore the driving force for aging of particular tissues or cells, and uncover potential intervention targets for aging and aging related diseases. Our recent related work includes, exploring regulatory network for epigenetic homeostasis in human mesenchymal stem cells (hMSC), revealing a set of rejuvenation factors that interact with nuclear envelope or heterochromatin proteins to stabilize heterochromatin and maintain the nuclear structure. These findings provide potential targets for intervention in aging or aging-related disorders, such as osteoarthritis and liver fibrosis (Figure 2). 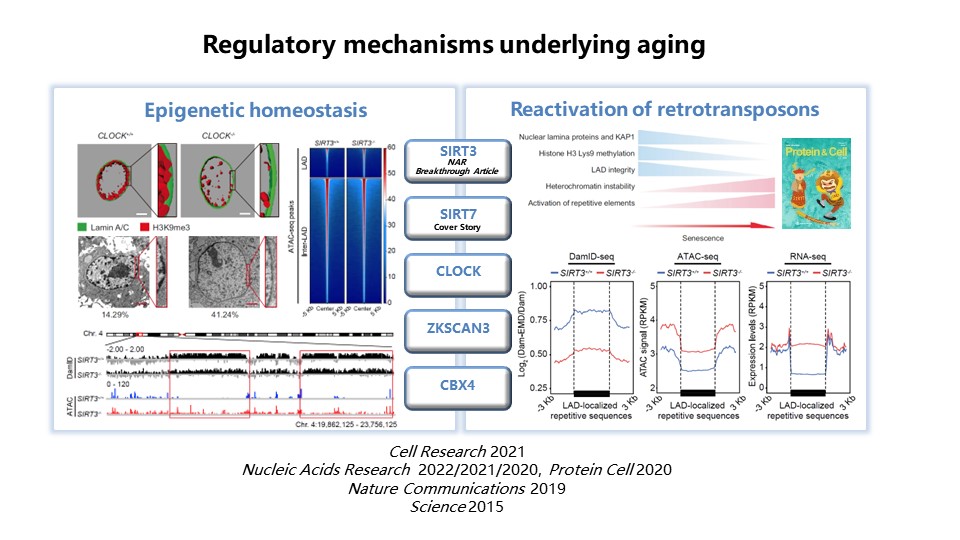
Figure 2. Systematic investigation of regulatory mechanisms underlying stem cell aging
Identifying a common molecular feature for primate arterial vascular cells, silencing of FOXO3A, and further demonstrating that FOXO3A silence is a driving force for arterial aging (Figure 3). 
Figure 3. Systematic investigation of regulatory mechanisms underlying organ/tissue aging
3) Establishment of intervention strategies for aging and aging-related diseases Using high-throughput screening platforms to discover rejuvenation factors that delay aging and promote regeneration, establishing novel intervention strategies against aging and related diseases based on gene editing and delivery techniques. Based on the in-depth understanding of aging, conferring stem cells with better ability to adapt to the aging microenvironment and resist malignant transformation. Our recent related work includes, editing specific nucleotides in the human genome to obtain the genetically enhanced stem cells and vascular cells; Discover a series of rejuvenation factors for gene therapy against osteoarthritis; A genome-wide CRISPR-based screen identifies KAT7 as a driver of cellular senescence. Lentiviral vectors encoding Cas9/sg-Kat7, given intravenously, alleviated hepatocyte senescence and liver aging and extended life span in physiologically aged mice as well as progeroid mice (Figure 4). 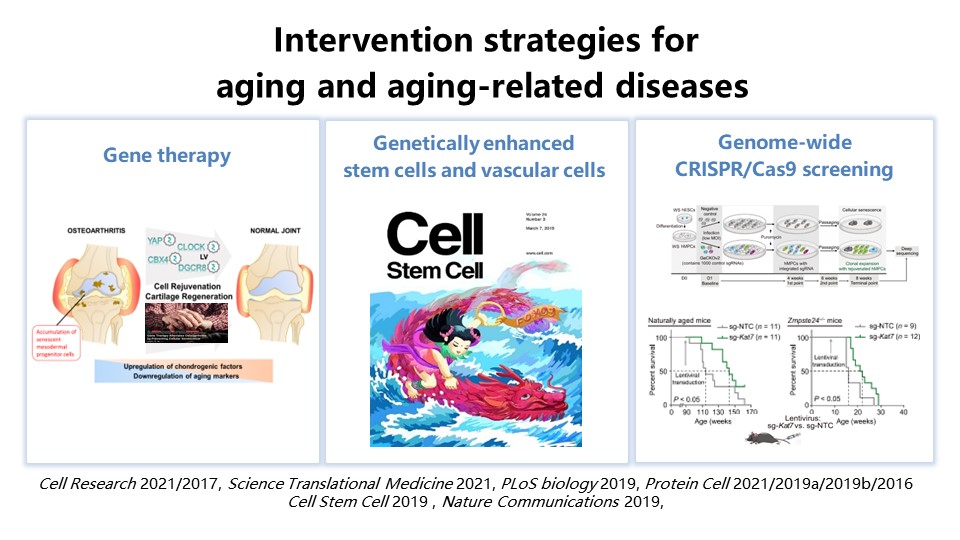
Figure 4. Intervention strategies for aging and aging-related diseases
| Selected publications: - The Sirtuin-associated human senescence program converges on the activation of placenta-specific gene PAPPA. Developmental Cell. 2024.
- Genome-wide CRISPR activation screening in senescent cells reveals SOX5 as a driver and therapeutic target of rejuvenation. Cell Stem Cell. 2023.
- Longevity secret - a pluripotent superpower. Cell Metabolism. 2022.
- Destabilizing heterochromatin by APOE mediates senescence. Nature Aging. 2022.
- A Single-cell Transcriptomic Atlas of Primate Pancreatic Islet Aging. National Science Review. 2021.
- SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Research. 2021.
- A Single-Cell Transcriptomic landscape of Primate Arterial Aging. Nature Communications. 2020.
- Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell. 2020. (Cover story)
- Caloric Restriction Reprograms the Single-Cell Transcriptional Landscape of Rattus Norvegicus Aging. Cell. 2020.
|

