| Current Research and Future Directions 1. Developing novel tools for genome engineering. TALEN andCRISPR systems were both discovered in microorganisms. Many enzymes essential for molecular biology, such as the restriction endonucleases, were discovered in microorganisms as well. Thanks to the development of genomics of microbiota and environmental microorganisms, millions of novel genes were discovered, the functions of which remains largely unknown. We are developing bioinformatic methods to identify candidate genes with potential of being novel genome engineering tools. We will screen these enzymes using in vitro biochemical assays and in vivo reporter assay in a high throughput manner, aiming for establishing novel tools that can be used to engineer the genomeof mammalian cells. 2. Establishment of efficient genome editing platform for therapeutic applications. Adoptive cell therapy such as chimeric antigen receptor (CAR) T cell therapy has been applied successfully in treating patients with various hematopoietic malignancies. However, many challenges remain in extending its application toward the treatment of solid tumors. We are aiming to develop more potent CAR-T technologyby applying gene editing to cell therapy. We are also developing high throughput genetic and chemical screening methods for human primary T cells, aiming to identify new targets and chemicals for immunotherapy. 3. Study the mechanism of X-chromosome inactivation (XCI) in human using stem cell and CRISPR technologies. To achieve gene dosage compensation between XX and XY individuals,X-chromosome inactivation (XCI) is evolved in mammals. The process is triggered by the long noncoding RNA Xist, which is expressed from the X-inactivation center (Xic). Although the mechanism of XCI has been studied in great details using mouse as in vivo model and mouse embryonic stem cells as in vitro models, the XCI process in human remains poorly understood due to the lack of proper models. By optimizing naive human embryonic stem cells culture and differentiation protocols, we achieved human random XCI in vitro.Using this platform, we will perform genetic screen using CRISPR technology to dissect the mechanism of XCI in human. 
Fig.1 TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors
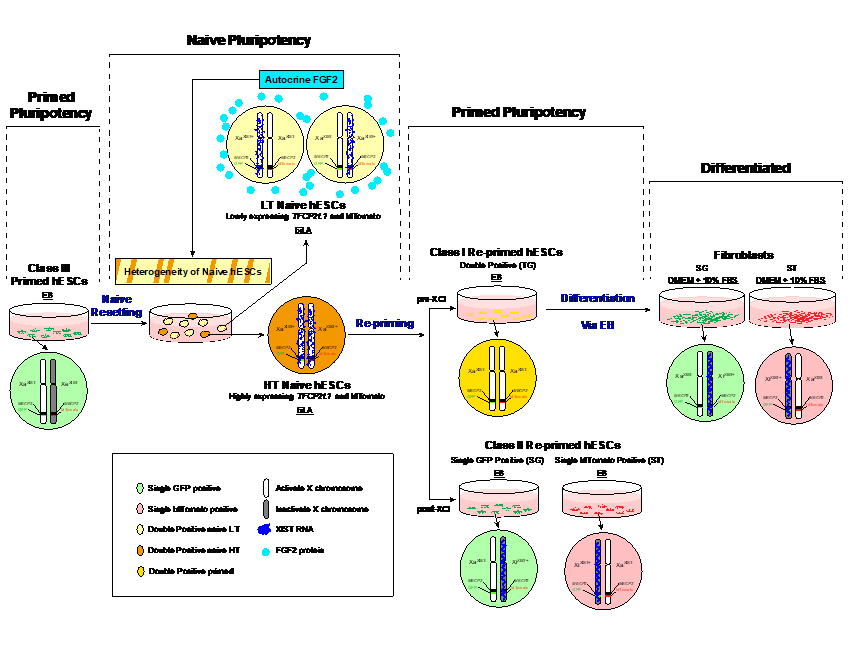
Fig.2 Modeling Human Random XCI Using Naive hESCs
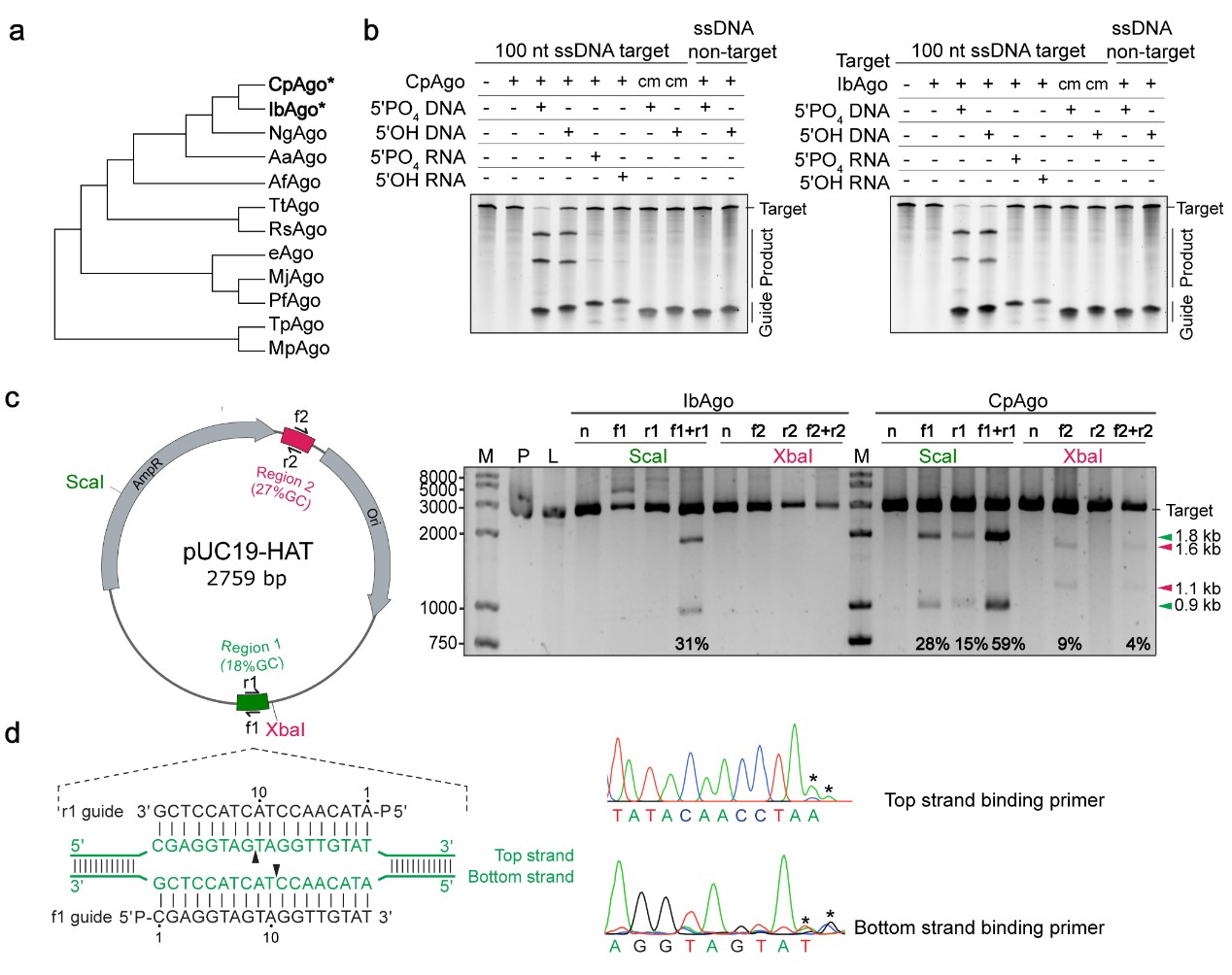
Fig.3 Characterization of the DNA cleavage activity of CpAgo and IbAgo
| Plain english:
Genome engineering technologies are invaluable tools for understanding the function of genes in development disease. In recent years, the programmable site-specific DNA endonucleases, including zinc finger nucleases (ZFNs), transcription activator–like effector nucleases (TALENs) and the clustered, regularly interspaced short palindromic–repeat (CRISPR) system, have gained tremendous popularity and become widely used for genome engineering in cell lines and animal species ranging from Drosophila to primates. Our laboratory focuses on developing novel technologies to achieve efficient and specific genome engineering, and applying them to study the function of genes and establish novel therapeutic methods.We currently have three main research interests:
1. Developing novel tools for genome engineering
2. Establishment of novel therapeutic methods using gene editing
3. Study the mechanism of X inactivation in human using stem cell and CRISPR technologies Selected publications:
(*Co-first author, # Corresponding author) - An C*, Feng G*, Zhang J, Cao S, Wang Y, Wang N, Lu F, Zhou Q, Wang H#. “Overcoming Autocrine FGF Signaling-Induced Heterogeneity in Naive Human ESCs Enables Modeling of Random X Chromosome Inactivation” Cell Stem Cell, 2020 Sep;27(3):482–497.
- Tang N*, Cheng C*, Zhang X, Qiao M, Li N, Mu W, Wei X, Han W, Wang H#. “TGFβ Inhibition via CRISPR Promotes the Long-Term Efficacy of CAR-T Cells Against Solid Tumors” JCI Insight, 2020 Feb;5(4): e133977.
- Cao Y*, Sun W*, Wang J, Sheng G, Xiang G, Zhang T, Shi W, Li C, Wang Y, Zhao F#, Wang H#. “Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double- stranded DNA at 37 °C.” Cell Discovery, 2019 Jul 30;5:38.
- Wang H#,Yang H#. “Gene-edited babies: What went wrong and what could go wrong.” PLOS Biology, 2019 Apr 30;17(4):e3000224.
- Mu W*, Tang N*, Cheng C*, Sun W, Wei X, Wang H#. “In vitro transcribed sgRNA causes cell death by inducing interferon release.” Protein & Cell,2019 Jun;10(6):461-465.
- Cheng C*, Tang N*, Li J, Cao S, Zhang T, Wei X, Wang H#. “ Bacteria-free minicircle DNA system to generate integration-free CAR-T cells.” Journal of Medical Genetics, 2019 Jan;56(1):10-17.
- Xiang G*, Ren J*, Hai T*, Fu R, Yu D, Wang J, Li W, Wang H#, Zhou Q#. “Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs.” Cellular and Molecular Life Sciences. 2018 Dec;75(24):4619-4628.
- Xiang G, Wang H#. “Extended pluripotent stem cells facilitate mouse model generation.” Protein & Cell. 2018 Aug 21.doi: 10.1007/s13238-018-0573-0.
- Mu W, Zhang Y, Xue X, Liu L, Wei X, Wang H#.” 5′ capped and 3′ polyA-tailed sgRNAs enhance the efficiency of CRISPR-Cas9 system.” Protein & Cell, 2018.Jun;10(3):223–228.
- Zhang Y, Mu W, Wang H#. “Gene editing in T cell therapy.” Journal of Genetics and Genomics, 2017 Sep;44 (9) :415-422.
- Wang H#. “Editing Base in Mouse Model.” Protein & Cell, 2017 Aug;8(8):558-559.
- Wang W, Zhang Y, Wang H#. “Generating Mouse Models Using Zygote Electroporation of Nucleases (ZEN) Technology with High Efficiency and Throughput.” Methods in Molecular Biology, 2017;1605:219-230.
- Zhang Y, Zhang X, Cheng C, Mu W, Liu X, Li N, Wei X, Liu X, Xia C, Wang H#. “CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells.” Frontiers of Medicine, 2017 Dec;11(4):554-562.
- Xiang G, Zhang X, An C, Cheng C, Wang H#. “Temperature effect on CRISPR-Cas9 mediated genome editing.” Journal of Genetics and Genomics, 2017 Apr ;44(4):199-205.
- Liu X, Zhang Y, Cheng C, Cheng WA, Zhang X, Li N, Xia C, Wei X, Liu X, Wang H#. “CRISPR-Cas9 mediated multiplex gene editing in CAR-T cells.” Cell Research, 2017 Jan;27(1):154-157.
- Burgess S, Cheng L#, Gu F, Huang J, Huang Z, Lin S#, Li J, Li W, Qin W, Sun W, Songyang Z, Wei W#, Wu Q, Wang H#, Wang X, Xiong JW, Xi J, Yang H, Zhou B, Zhang B. “Questions about NgAgo” Protein Cell, 2016 Dec;7(12):913-915.
- Wang W,Kutny PM, Byers SL, Longstaff CJ, DaCosta MJ, Pang C, Zhang Y, Taft RA, Buaas FW, Wang H#. “Delivery of Cas9 protein into mouse zygotes through a series of electroporation dramatically increased the efficiency of model creation.” Journal of Genetics and Genomics, 2016 May 20;43(5):319-27.
- Qin W, Kutny PM, Maser RS, Dion SL, Lamont JD, Zhang Y, Perry GA, Wang H#.“Generating Mouse Models Using CRISPR-Cas9 Mediated Genome Editing.”Current Protocols in Mouse Biology, 2016 Mar 1;6(1):39-66.
- Cheng AW*#, Jillette N*, Lee P, Plaskon D, Fujiwara Y, Wang W, Taghbalout A, Wang H#. “Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling.” Cell Research, 2016 Feb;26(2):254-7.
- Wiles MV, Qin W, Cheng AW, Wang H#. “CRISPR-Cas9 mediated genome editing and guide RNA design.” Mammalian Genome, 2015 Oct;26(9-10):501-10.
- Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, Malhotra A, Geurts AM, Chen YG, Wang H#. “Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease.” Genetics, 2015 Jun;200(2):423-30.
- Theunissen T*, Powell B*, Wang H*, Mitalipova M, Faddah D, Reddy J, Fan Z, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D’Alessio A, Zhang J, Gao Q, Dawlaty M, Young R, Gray N, Jaenisch R. “Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency.”Cell Stem Cell, 2014 Oct 2; 15(4):471-87.
- Yang H, Wang H, Jaenisch R. “Generating genetically modified mice using CRISPR/Cas-mediated genome engineering.” Nature protocols, 2014 Aug; 9(8):1956-68.
- Maetzel D*,Sarkar S*, Wang H*, Mosleh LA, Cheng AW, Xu P, Gao Q, Mitalipova M, Jaenisch R. “Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells.” Stem Cell Reports, 2014 May 15; 2(6):866-80.
- Cheng AW*, Wang H*, Yang H, Shi L, Katz Y, Rangarajan S, Theunissen TW, Shivalila CS, Dadon DB, Jaenisch R. “Multiplexed activation of endogenous genes by CRISPR-on, a RNA-guided transcriptional activator system.” Cell Research, 2013 October 1; 23(10): 1163-1171.
- Yang H*, Wang H*, Shivalila CS*, Cheng AW, Shi L, Jaenisch R. “One-step generation of mice carrying reporter and conditional allele by CRISPR/Cas mediated genome editing” Cell, 2013 September 12; 154(6):1370-1379.
- Faddah D, Wang H, Buganim Y, Cheng AW, Jaenisch R. “Expression of Nanog is biallelic and equally variable as other pluripotency factors.” Cell Stem Cell, 2013 Jul 3;13(1):23-9.
- Wang H*, Hu YC*, Markoulaki S, Welstead GG, Shivalila CS, Cheng AW, Pyntikova T, Dadon D, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R. “TALEN-mediated editing of the Mouse Y Chromosome.” Nature Biotechnology, 2013 Jun; 31(6):530-2.
- Wang H*, Yang H*, Shivalila CS*, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. “One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas mediated genome engineering.” Cell, 2013 May 9;153(4):910-8.
|

