 | Wei LI, Ph.D., Professor Genetic engineering, cell/animal models
Staff: Wei LI, Tianda LI, Zhikun LI, Zhengquan HE
Postdoc:Tongtong CUI, Chao LIU, Jingjing LIU, Yihuan MAO, Fei TENE, Yanping HU, Tingting GAO, Jilong REN
Students: Moyu DAI, Weiqi GUO, Peiqi LI, Najmeh Karimi, Rongqi LI, Chen LIANG, Shasha LIU, Bangwei MAO, Xuehan SUN, Lin WANG, Jing XU, Ning YANG, Jin ZHANG, Yulong ZHAO, Li ZHOU, Liyu ZHU
| | | | | | An old but still unaccomplished goal in biology is to elucidate which and how, genes and their regulations, determine a phenotype/function. Although a series of landmark achievements have emerged in answering this question, it is still very challenging to reconstruct a phenotype/function completely using designed genetic modules. The long-term goal of my research is to understand the genetic basis of certain reproduction- and regeneration-related functions/phenotypes, and to reconstitute such phenotype/function in a given cell or organism for research and therapeutic purposes. Current interests in my laboratory include: (i) developing cutting-edge genome engineering technologies and mammalian synthetic biology. (ii) dissection of genetic and epigenetic basis of reproduction- and regeneration-related phenotypes. (iii) clinical application of gene therapy. 
One adult mouse on the bamboo that carry a ligated chromosome by fusing chromosome 4 and 5
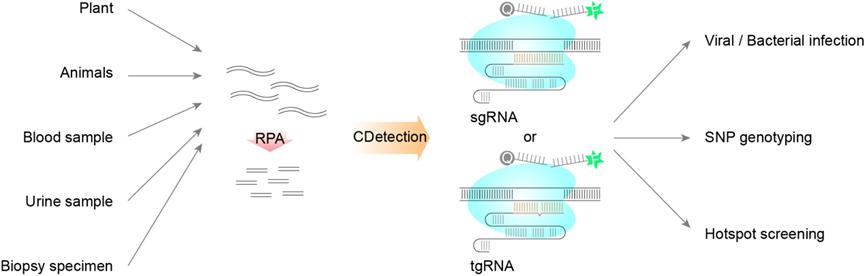
CDetection has high sensitivity and accuracy, and provides an efficient and highly practical platform for DNA detection.
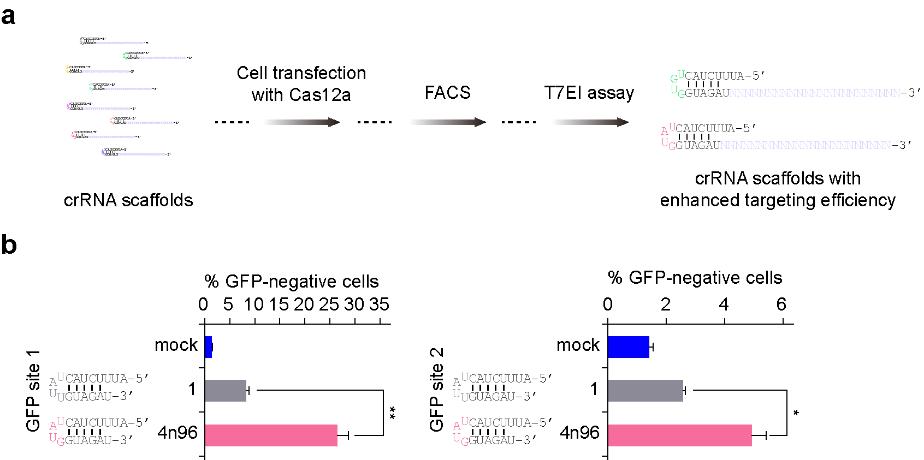
Enhanced targeted indel frequencies by optimized crRNA scaffold. a Schematic showing the workflow of crRNA scaffold optimization and screening. b Efficiencies of GFP disruption in human 293FT cells generated by PrCas12a/scaffold 1 and PrCas12a/scaffold 4n96, respectively. Error bars indicate standard errors of the mean (SEM), n = 3. *p value < 0.05 and **p value < 0.01.
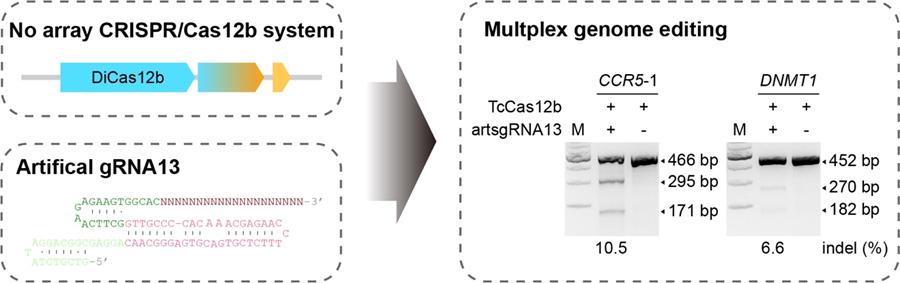
Artificial sgRNAs engineered for genome editing with new Cas12b orthologs
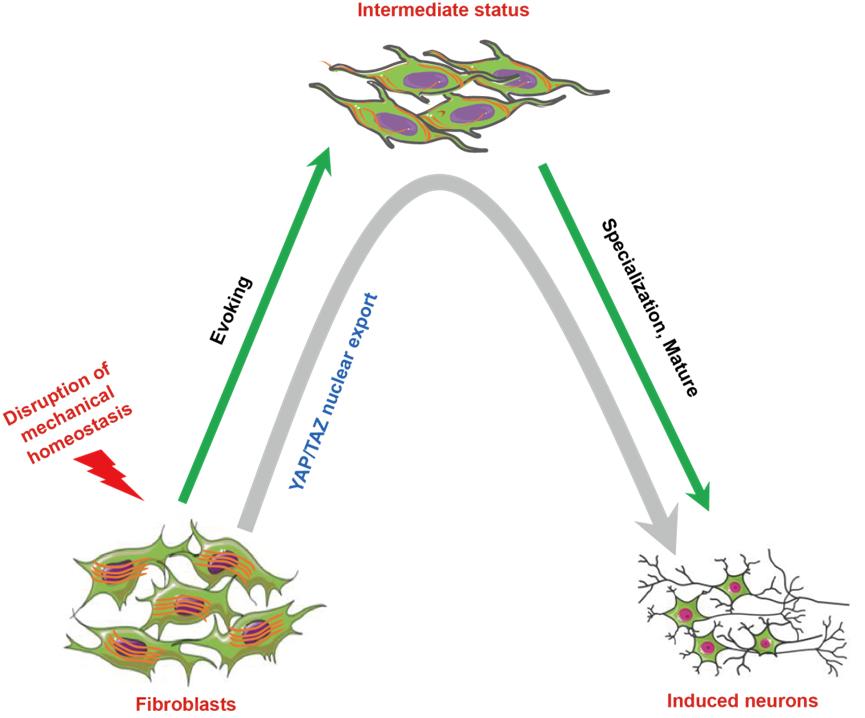
Model of neuronal conversion mediated through the disruption of mechanical homeostasis in fibroblasts.
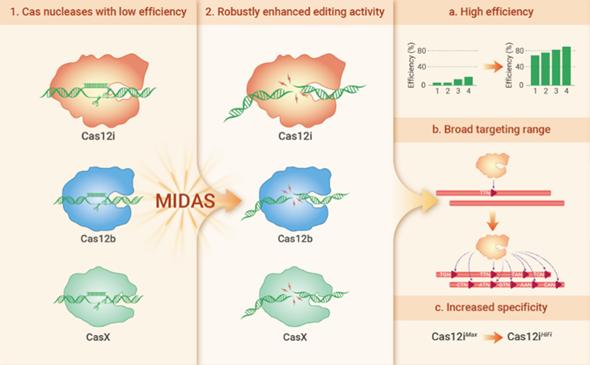
Synergistic engineering (MIDAS) of CRISPR-Cas nucleases obtains high-efficiency Cas12iMax and high-specificity Cas12iHiFi.
| Plain English:
Current interests in my laboratory include: (i) developing genome engineering cutting-edge genome engineering technologies and mammalian synthetic biology. (ii) dissection of genetic and epigenetic basis of reproduction- and regeneration-related phenotypes. (iii) clinical application of gene therapy. Selected publications: - Cui T#, Cai B#, Tian Y, Liu X, Liang C, Gao Q, Li B, Ding Y, Li R, Zhou Q, Li W*, Teng F*. Therapeutic In Vivo Gene Editing Achieved by a Hypercompact CRISPR-Cas12f1 System Delivered with All-in-One Adeno-Associated Virus. Advanced Science (Weinh) 2024 Feb 26:e2308095. doi: 10.1002/advs.202308095. Online ahead of print.
- Wang C#, Fang S#, Chen Y#, Tang N, Jiao G, Hu Y, Li J, Shan Q, Wang X, Feng G, Zhou Q*, Li W*. High-efficiency targeted transgene integration via primed micro-homologues. Cell Discovery 2023 Jul 4;9(1):69. doi: 10.1038/s41421-023-00552-0.
- Wang LB#, Li ZK#, Wang LY#, Xu K#, Ji TT#, Mao YH, Ma SN, Liu T, Tu CF, Zhao Q, Fan XN, Liu C, Wang LY, Shu YJ, Yang N, Zhou Q*, Li W*. A sustainable mouse karyotype created by programmed chromosome fusion. Science 2022 Aug 26;377(6609):967-975. doi: 10.1126/science.abm1964. Epub 2022 Aug 25.
- He ZQ#, Li YH#, Feng GH#, Yuan XW#, Lu ZB#, Dai M, Hu YP, Zhang Y, Zhou Q*, Li W*. Pharmacological Perturbation of Mechanical Contractility Enables Robust Transdifferentiation of Human Fibroblasts into Neurons. Advanced Science (Weinh). 2022 May;9(13):e2104682. doi: 10.1002/advs.202104682.
- Chen Y#, Hu Y#, Wang X#, Luo S, Yang N, Chen Y, Li Z, Zhou Q*, Li W*. Synergistic engineering of CRISPR-Cas nucleases enables robust mammalian genome editing. Innovation (Camb). 2022 May 26;3(4):100264. doi: 10.1016/j.xinn.2022.100264.
- Guo L#, Sun X#, Wang X#, Liang C#, Jiang H#, Gao Q#, Dai M#, Qu B, Fang S, Mao Y, Chen Y, Feng G, Gu Q, Wang RR*, Zhou Q*, Li W*. SARS-CoV-2 detection with CRISPR diagnostics. (2020) Cell Discovery 6:34. doi: 10.1038/s41421-020-0174-y.
- Wang LY#, Li ZK#, Wang LB#, Liu C#, Sun XH, Feng GH, Wang JQ, Li YF, Qiao LY, Nie H, Jiang LY, Sun H, Xie YL, Ma SN, Wan HF, Lu FL*, Li W*, Zhou Q*. Overcoming Intrinsic H3K27me3 Imprinting Barriers Improves Post-implantation Development after Somatic Cell Nuclear Transfer. (2020) Cell Stem Cell 27(2):315-325.e5. doi: 10.1016/j.stem.2020.05.014.
- Qu B#, Wu S#, Jiao G#, Zou X#, Li Z#, Guo L#, Sun X, Huang C, Sun Z, Zhang Y, Li H, Zhou Q, Sui R*, Li W*. Treating Bietti crystalline dystrophy in a high-fat diet-exacerbated murine model using gene therapy. (2020) Gene Therapy 27(7-8):370-382.doi: 10.1038/s41434-020-0159-3.
- Teng F#, Guo L#, Cui T, Wang XG, Xu K, Gao Q, Zhou Q*, Li W*. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. (2019) Genome Biology 20(1):132. doi: 10.1186/s13059-019-1742-z.
- Teng F#, Li J#, Cui T#, Xu K, Guo L, Gao Q, Feng G, Chen C, Han D, Zhou Q*, Li W*. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. (2019) Genome Biology 20(1):15. doi: 10.1186/s13059-019-1620-8.
- Wang J#, Wang L#, Feng G#, Wang Y#, Li Y, Li X, Liu C, Jiao G, Huang C, Shi J, Zhou T, Chen Q, Liu Z, Li W*, Zhou Q*. Asymmetric Expression of LincGET Biases Cell Fate in Two-Cell Mouse Embryos. (2018) Cell 175(7):1887-1901.e18. doi: 10.1016/j.cell.2018.11.039.
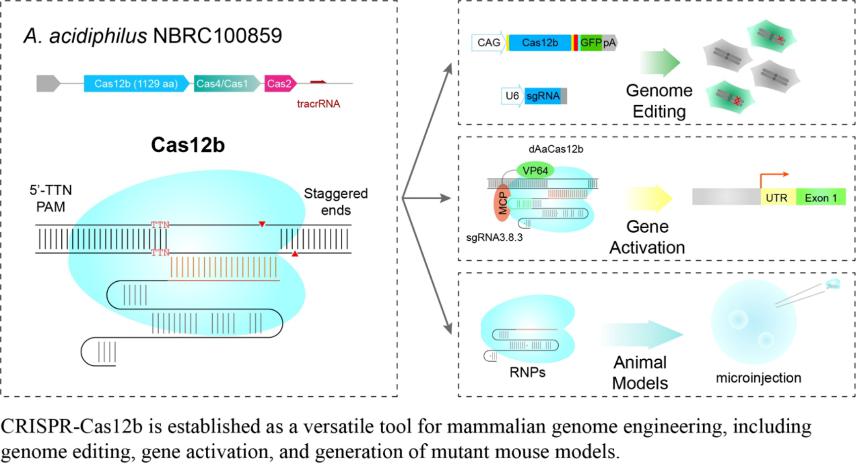
- Teng F#, Cui T#, Feng G, Guo L, Xu K, Gao Q, Li T, Li J, Zhou Q, Li W*. Repurposing CRISPR-Cas12b for mammalian genome engineering. (2018) Cell Discovery 4:63. doi: 10.1038/s41421-018-0069-3.
- Zhang W#, Wan H#, Feng G#, Qu J#, Wang J, Jing Y, Ren R, Liu Z, Zhang L, Chen Z, Wang S, Zhao Y, Wang Z, Yuan Y, Zhou Q, Li W*, Liu G*, Hu B*. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. (2018) Nature 560(7720):661-665. doi: 10.1038/s41586-018-0437-z.
- Li ZK#, Wang LY#, Wang LB#, Feng GH#, Yuan XW#, Liu C, Xu K, Li YH, Wan HF, Zhang Y, Li YF, Li X, Li W*, Zhou Q*, Hu BY*. Generation of Bimaternal and Bipaternal Mice from Hypomethylated Haploid ESCs with Imprinting Region Deletions. (2018) Cell Stem Cell 23(5):665-676.e4. doi: 10.1016/j.stem.2018.09.004.
- Xu K#, Yang Y#, Feng GH#, Sun BF#, Chen JQ#, Li YF, Chen YS, Zhang XX, Wang CX, Jiang LY, Liu C, Zhang ZY, Wang XJ, Zhou Q*, Yang YG*, Li W*. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. (2017) Cell Research 27(9):1100-1114. doi: 10.1038/cr.2017.100. (Cover story)
- Li X#, Cui XL#, Wang JQ#, Wang YK, Li YF, Wang LY, Wan HF, Li TD, Feng GH, Shuai L, Li ZK, Gu Q, Hao J, Wang L, Zhao XY, Liu ZH, Wang XJ, Li W*, Zhou Q*. Generation and application of mouse-rat allodiploid embryonic stem cells. (2016) Cell 164(1-2):279-292. doi: 10.1016/j.cell.2015.11.035.
- Wan HF#, Feng CJ#, Teng F#, Yang SH#, Hu BY, Niu YY, Xiang P, Fang WZ, Ji WZ, Li W*, Zhao XY*, Zhou Q*. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. (2015) Cell Research 25(2):258-261. doi: 10.1038/cr.2014.158.
- Li W#, Li X#, Li TD#, Jiang MG#, Wan HF, Luo GZ, Feng CJ, Cui XL, Teng F, Yuan Y, Zhou Q, Gu Q, Shuai L, Sha JH, Xiao YM, Wang L, Liu ZH, Wang XJ, Zhao XY, Zhou Q*. Genetic modification and screening in rat using haploid embryonic stem cells. (2014) Cell Stem Cell 14(3):404-414. doi: 10.1016/j.stem.2013.11.016.
- Li W#, Teng F#, Li T, Zhou Q*. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. (2013) Nature Biotechnology 31(8):684-686. doi: 10.1038/nbt.2652.
- Li W#, Shuai L#, Wan H#, Dong M#, Wang M, Sang L, Feng C, Luo GZ, Li T, Li X, Wang L, Zheng QY, Sheng C, Wu HJ, Liu Z, Liu L, Wang L, Wang XJ, Zhao XY*, Zhou Q*. Androgenetic haploid embryonic stem cells produce live transgenic mice. (2012) Nature 490(7420):407-411. doi: 10.1038/nature11435.
| |

