| Pigs are playing important roles in biomedical research and serving as important sources of meat. Our lab is committed to building an efficient platform for pig genetic modifications and functional genomics analysis, providing ideal disease models for biomedical research and preclinical transformation, and providing new solutions for pig economic trait improvement and breeding. The main research directions are as follows: 1. Establishment of mutant lines and screening of functional genes by whole-genome random mutagenesis; 2. Genetic modifications by using various genome editing tools to achieve the agricultural and biomedical applications of pigs; 3. Reprogramming mechanism in somatic cell nuclear transfer and epigenetic regulations in the development of porcine early stage embryos. 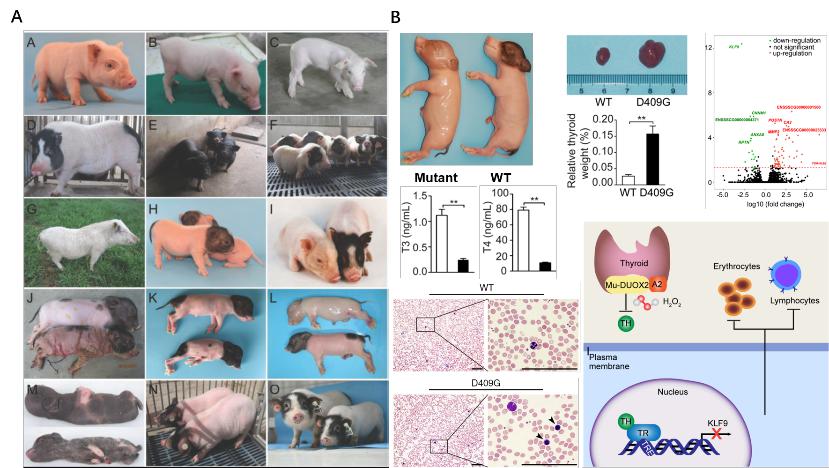
A large number of animal models were created using ENU mutagenesis, and a large animal model for human congenital hypothyroidism (CH) was generated and successfully analyzed.
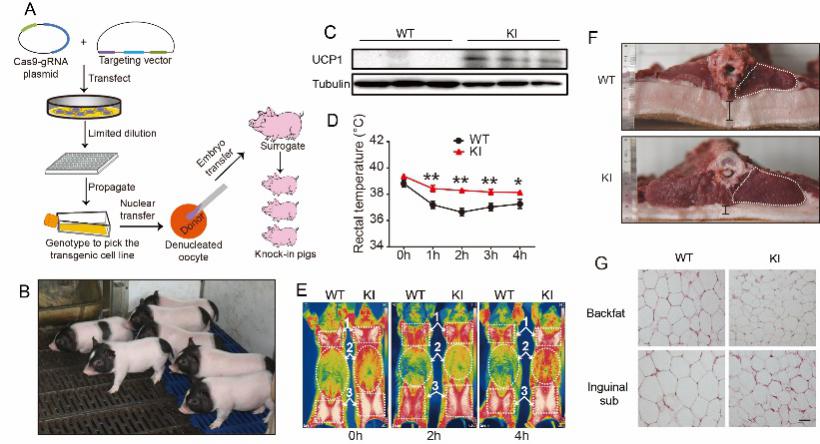
Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity.
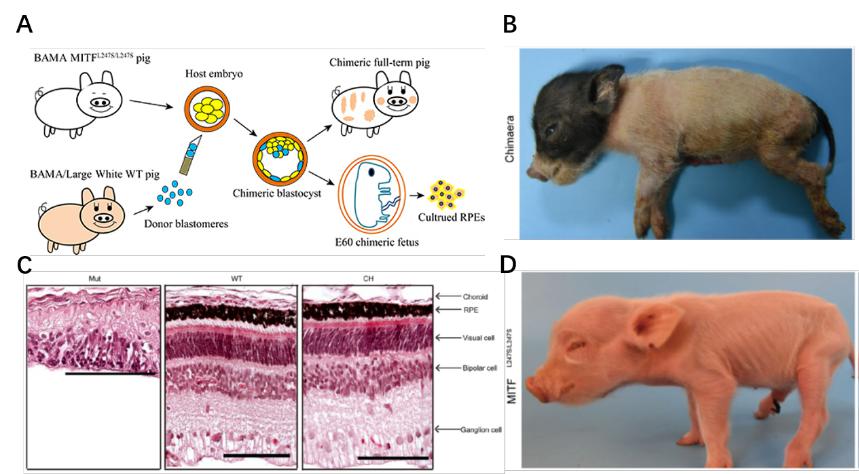
Rescuing ocular development in an anophthalmic pig by blastocyst complementation.
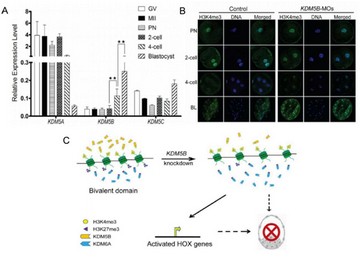
KDM5B is critical to porcine embryo development through regulating the balance of bivalent H3K4me3-H3K27me3 modifications.
| Plain english:
Pigs have been widely used in biomedical researches, such as human disease modeling, xeno-transplantation, and therapeutics. They are also essential in agriculture as important sources of meat. Our lab focuses on creation of human disease models and improvement of economic traits in pigs by using forward and reverse genetics approaches. Selected publications: - Wang X§, Cao C§, Zhang Y, Jia Q, Zheng Q, Zhang H, Song R, Li Y, Luo A, Hong Q, Qin G, Yao J, Zhang N, Wang Y, Zhou Q, Zhao JG*. A harlequin ichthyosis pig model with a novel ABCA12 mutation can be rescued by acitretin treatment. J Mol Cell Biol, 2019, 11 (12), 1029-1041. (Cover story)
- Zhao JG§, Lai L, Ji W, Zhou Q*. Genome editing in large animals: Current status and future prospects. Natl Sci Rev, 2019, 6: 402-420. (Invited review)
- Cao C§, Zhang Y§, Jia Q, Wang X, Zheng Q, Zhang H, Song R, Li Y, Luo A, Hong Q, Qin G, Yao J, Zhang N, Wang Y, Wang H,Zhou Q, Zhao JG*. An exonic splicing enhancer mutation in DUOX2 causes aberrant alternative splicing and severe congenital hypothyroidism in Bama pigs. Dis Model Mech, 2019, 12: dmm036616.
- Zhang Y§, Hong Q, Cao C, Yang L, Li Y, Hai T, Zhang H, Zhou Q, Sui R*, Zhao J*. A novel porcine model reproduces human oculocutaneous albinism type II. Cell discovery, 2019, 5: 48.
- Zhang H§, Huang J§, Li Z§, Qin G, Zhang N, Hai T, Hong Q, Zheng Q, Zhang Y, Song R, Yao J, Cao C, Zhao JG*, Zhou Q*. Rescuing ocular development in an anophthalmic pig by blastocyst complementation. EMBO Mol Med. 2018;10(12). pii: e8861.
- Wang Y§*, Huang JJ, Zhao JG*. Gene engineering in swine for agriculture. J Integr Agr. 2017; 16(12): 2792-2804.
- Hai T§, Guo W§, Yao J§, Cao C, Luo A, Qi M, Wang X, Wang X. Huang J, Zhang Y, Wang D, Shang H, Hong Q, Zhang R, Jia Q, Zheng Q, Qin G, Li Y, Zhang T, Jin W, Chen ZY, Wang H, Zhou Q, Meng A, Wei H*, Yang S*, Zhao JG*. Creation of miniature pig model of human Waardenburg syndrome type 2A by ENU mutagenesis. Human genetics. 2017; 136:1463-1475.
- Zheng Q§, Lin J§, Huang J§, Zhang H, Zhang R, Zhang X, Cao C, Hambly C, Qin G, Yao J, Song R, Jia Q, Wang X, Li Y, Zhang N, Piao Z, Ye R, Speakman JR, Wang H, Zhou Q, Wang Y*, Jin W*, Zhao JG*. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decrease fat deposition and improve thermogenic capacity. Proc Natl Acad Sci U S A. 2017; 114(45): E9474-E9482.
- Zhang Y§, Xue Y§, Cao C§,Huang J, Hong Q, Hai T, Jia Q, Wang X, Qin G, Yao J, Wang X, Zheng Q, Zhang R, Li Y, Luo A, Zhang N, Shi G, Wang Y, Ying H, Liu Z, Wang H, Meng A, Zhou Q, Wei H*, Liu F*, Zhao JG*. Thyroid hormone regulates hematopoiesis via the TR-KLF9 axis. Blood. 2017; 130(20):2161-2170.
- Huang J§, Wang Y, Zhao J*. CRISPR editing in biological and biomedical investigation. J Cell Physiol. 2018 May;233(5):3875-3891
- Hai T§, Cao C§, Shang H§, Guo W§, Mu Y§, Yang S, Zhang Y, Zheng Q, Zhang T, Wang X, Liu Y, Kong Q, Li K, Wang D, Qi M, Hong Q, Zhang R, Wang X, Jia Q, Wang X, Qin G, Li Y, Luo A, Jin W, Yao J, Huang J, Zhang H, Li M, Xie X, Zheng X, Guo K, Wang Q, Zhang S, Li L, Xie F, Zhang Y, Weng X, Yin Z, Hu K, Cong Y, Zheng P, Zou H, Xin L, Xia J, Ruan J, Li H, Zhao W, Yuan J, Liu Z, Gu W, Li M, Wang Y, Wang H*, Yang S*, Liu Z*, Wei H*, Zhao JG*, Zhou Q*, Meng A*. Pilot study of large-scale production of mutant pigs by ENU mutagenesis. Elife. 2017 Jun 22; 6. pii: e26248.
- Lin J§, Cao C§, Tao C§, Ye R, Dong M, Zheng Q, Wang C, Jiang X, Qin G, Yan C, Li K, Speakman JR, Wang Y*, Jin W*, Zhao JG*. Cold adaptation in pigs depends on UCP3 in beige adipocytes. J Mol Cell Biol. 2017 Oct 1; 9(5):364-375.
- Jia Q§, Cao C§, Tang H, Zhang Y, Zheng Q, Wang X, Zhang R, Wang X, Luo A, Wei H, Meng A, Zhou Q, Wang H*, Zhao JG*. A 2-bp insertion (c.67_68insCC) in MC1R causes recessive white coat color in Bama miniature pigs. J Genet Genomics. 2017 Apr 20; 44(4):215-217.
- Yao J§, Huang J, Zhao JG*. Genome editing revolutionize the creation of genetically modified pigs for modeling human diseases. Hum Genet. 2016 Sep;135(9):1093-105.
- Wang X§, Cao C§, Huang J§, Yao J, Hai T, Zheng Q, Wang X, Zhang H, Qin G, Cheng J, Wang Y, Yuan Z, Zhou Q, Wang H*, Zhao JG*. One-step generation of triple gene targeted pigs using CRISPR/Cas9 system. Sci Rep. 2016; 6:20620.
- Huang J§, Zhang H, Yao J, Qin G, Wang F, Wang X, Luo A, Zheng Q, Cao C, Zhao JG*. BIX-01294 increases pig cloning efficiency by improving epigenetic reprogramming of somatic cell nuclei. Reproduction. 2016 Jan; 151(1):39-49.
- Zhao JG§. , Xu WJ, Ross JW., Walters EM., Butler SP., Whyte JJ., Kelso L., Fatemi M, Vanderslice NC., Giroux K., Spate LD., Samuel MS., Murphy CN., Wells KD., Masiello NC., Prather RS*. & Velander WH*. Engineering protein processing of the mammary gland to produce abundant hemophilia B therapy in milk. Sci Rep. 2015 Sep 21; 5: 14176.
- Wang X§, Zhou J§, Cao C§, Huang J, Hai T, Wang Y, Zheng Q, Zhang H, Qin G, Miao X, Wang H, Cao S*, Zhou Q*, Zhao JG*. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci Rep. 2015 Aug 21; 5: 13348.
- Zhao W§, Wang Y§, Liu S, Huang J, Zhai Z, He C, Ding J, Wang J, Wang H, Fan W, Zhao JG*, Meng H*.The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One. 2015, 10(2): e0117441.
- Huang J§, Zhang H, Wang X, Dobbs KB, Yao J, Qin G, Whitworth K, Walters EM, Prather RS*, Zhao JG*. Impairment of Preimplantation Porcine Embryo Development by Histone Demethylase KDM5B Knockdown Through Disturbance of Bivalent H3K4me3-H3K27me3 Modifications. Biol Reprod. 2015, 92(3):72. (*Co-corresponding Author)
- Yao J§, Huang JJ, HaiTang , Wang XL, Qin GS, Zhang HY, Wu R, Xi J. J, Yuan Z, Zhao JG*. Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Sci Rep. 2014, 4:6926. (*Corresponding Author)
- Wang YP§, Qi ST, Wei Y, Ge ZJ, Chen L, Hou Y, Ouyang YC, Schatten H, Zhao JG*, Sun QY*.Knockdown of UCHL5IP causes abnormalities in γ-tubulin localisation, spindle organisation and chromosome alignment in mouse oocyte meiotic maturation. Reprod Fertil Dev. 2013; 25(3):495-502. (*Co-corresponding Author)
- Whyte JJ§,Zhao JG§, Samuel M, Laughlin HM, Prather RS*, Gene Targeting with Zinc Finger Nucleases to Produce Cloned eGFP Knockout Pigs. Mol Reprod Dev. 2011, 78(1):2.
- Zhao JG§, Whyte JJ, Prather RS*. The effect of epigenetic regulation on somatic cell nuclear transfer. Cell. Cell Tissue Res. 2010, 341(1):13-21.
- Zhao JG§, Hao Y, Ross JW, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS*. Histone Deacetylase Inhibitors Improve In Vitro and In Vivo Developmental Competence of Somatic cell Nuclear Transfer Porcine Embryos.Cloning and stem cells,2010,12(1):75-83.
- Ross JW§, Prather RS, Whyte JJ,Zhao JG*.Cloning and transgenics: progress and new approaches.Invited review by Centre for Agricultural Bioscience International,2009, 4, No. 38.
- Zhao J§, Ross JW, Hao Y, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS*. Significant Improvement in Cloning Efficiency of an Inbred Miniature Pig by Histone Deacetylase Inhibitor Treatment after Somatic Cell Nuclear Transfer. Biol Reprod. 2009, 81: 525-530.

|

